A review on CRISPR/Cas9 system for the research and control of helminth parasites
2020-11-20 11:51
Globally, parasitic helminths afflict billions of people and die of resulting complications every year. Currently, no effective vaccines are available for human use against any of these parasitic worms. The limited chemotherapy options for treatment of the diseases caused by parasitic helminths increase the risk that drug resistance will develop. It is, thus, important that improved control strategies be developed using advanced techniques that can mitigate this 21st-century threat to global health through the identification of novel drugs or vaccine intervention targets.
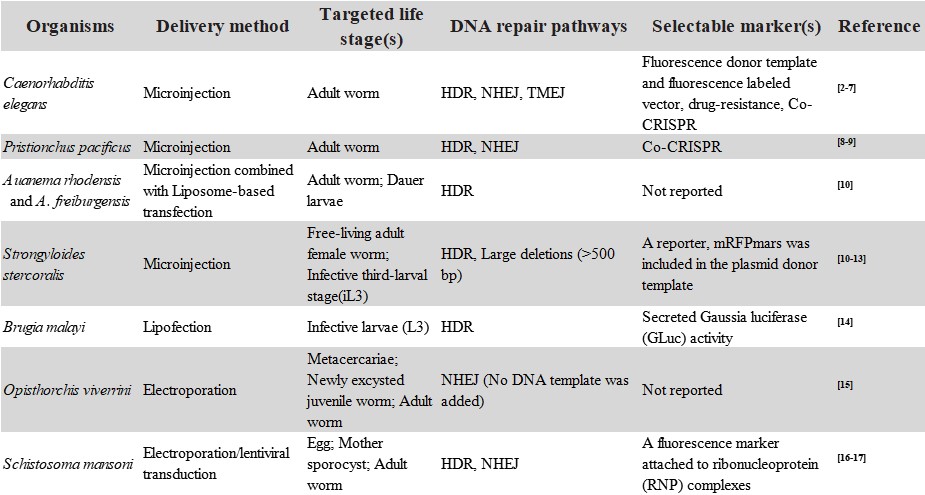
Recent reports of CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein form adaptive immunity system of bacteria and archaea to prevent bacteriophage) gene editing technology in parasitic helminths open up new avenues for research on these dangerous pathogens. It has been successfully established in four free-living nematodes, three parasitic nematodes and two parasitic trematodes (Table 1).
However, the complex morphology and life cycles inherent to these parasites present obstacles for the efficient application of CRISPR/Cas9-targeted mutagenesis. This is especially true with the trematode flukes where only modest levels of gene mutation efficiency have been achieved. Major challenges in the application of CRISPR/Cas9 for study of parasitic worms thus lie in enhancing gene mutation efficiency and overcoming issues involved in host passage so that mutated parasites survive, and developing strategies of gene editing using CRISPR/Cas9 system on them, for example, novel delivery methods, the choice of selectable markers and refining mutation precision represent novel tactics. Recently, Du X et al. review and summarize information of helminth parasites genome editing [1].
Table 1 Use of CRISPR/Cas9 editing system in helminths
References:
[1] Du X, McManus DP, French JD, Jones MK, You H. CRISPR/Cas9: A new tool for the study and control of helminth parasites. Bioessays. 2020;e2000185. doi:10.1002/bies.202000185
[2] Friedland, A. E., Tzur, Y. B., Esvelt, K. M., Colaiácovo, M. P., Church, G.M., & Calarco, J. A. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods, 10(8), 741–749.
[3] Van Schendel, R., Roerink, S. F., Portegijs, V., Van Den Heuvel, S., & Tijsterman, M. (2015). Polymerase Θ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun., 6, 7394.
[4] Chen, C., Fenk, L. A., & de Bono, M. (2013). Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res., 41(20), e193.
[5] Dickinson, D. J., Ward, J. D., Reiner, D. J., & Goldstein, B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods, 10(10), 1028–1034.
[6] Paix, A., Folkmann, A., Rasoloson, D., & Seydoux, G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics, 201(1), 47–54.
[7] Arribere, J. A., Bell, R. T., Fu, B. X., Artiles, K. L., Hartman, P. S., & Fire, A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics, 198(3), 837–546.
[8] K.-i. Nakayama, Ishita, Y., Chihara, T., & Okumura, M. (2020). Screening for CRISPR/Cas9-induced mutations using a co-injection marker in the nematode Pristionchus pacificus. Dev. Genes Evol., 1, 257–264.
[9] Witte, H., Moreno, E., Rödelsperger, C., Kim, J., Kim, J. - S., Streit, A., & Sommer, R. J. (2015). Gene inactivation using the CRISPR/Cas9 system in the nematode Pristionchus pacificus. Dev. Genes Evol., 225(1), 55–62.
[10] Adams, S., Pathak, P., Shao, H., Lok, J. B., & Pires-daSilva, A. (2019). Liposome-based transfection enhances RNAi and CRISPR-mediated mutagenesis in non-model nematode systems. Sci. Rep., 9(1), 483.
[11] Gang, S. S., Castelletto, M. L., Bryant, A. S., Yang, E., Mancuso, N., Lopez, J. B., . . . Hallem, E. A. (2017). Targeted mutagenesis in a human[1]parasitic nematode. PLoS Pathog., 13(10), e1006675.
[12] Bryant, A. S., Ruiz, F., Gang, S. S., Castelletto, M. L., Lopez, J. B., & Hallem, E. A. (2018). A critical role for thermosensation in host seeking by skin-penetrating nematodes. Curr. Biol., 28(14), 2338– 2347.
[13] Lok, J., Shao, H., Massey, H., & Li, X. (2017). Transgenesis in Strongyloides and related parasitic nematodes: Historical perspectives, current functional genomic applications and progress towards gene disruption and editing. Parasitology, 144(3), 327–342.
[14] Liu, C., Grote, A., Ghedin, E., & Unnasch, T. R. (2020). CRISPR[1]mediated transfection of Brugia malayi. PLoS Negl. Trop. Dis., 14(8), e0008627.
[15] Arunsan, P., Ittiprasert, W., Smout, M. J., Cochran, C. J., Mann, V. H., Chaiyadet, S., . . . Sotillo, J. (2019). Programmed knockout mutation of liver fluke granulin attenuates virulence of infection-induced hepato-biliary morbidity. Elife, 8, e41463.
[16] Ittiprasert, W., Mann, V. H., Karinshak, S. E., Coghlan, A., Rinaldi, G., Sankaranarayanan, G., . . . Prangtaworn, P. (2019). Programmed genome editing of the omega-1 ribonuclease of the blood fluke, Schistosoma Mansoni. Elife, 8, e41337.
[17] You, H., Mayer, J. U., Johnston, R. L., Sivakumaran, H., Ranasinghe, S., Rivera, V., . . . Driguez, P. (2020). CRISPR/Cas9-mediated genome editing of Schistosoma mansoni acetylcholinesterase. bioRxiv.
[18] Sankaranarayanan, G., Coghlan, A., Driguez, P., Lotkowska, M. E., Sanders, M., Holroyd, N., . . . Rinaldi, G. (2020). Large CRISPR-Cas[1]induced deletions in the oxamniquine resistance locus of the human parasite Schistosoma mansoni. Wellcome Open Res., 5, 178.



