Effects of ethanol exposure on the transcriptome of C. elegans
2021-07-20 10:45
Alcohol is commonly used in modern life. Moderate drinking has the function of promoting blood circulation and preventing cardiovascular diseases. However, excessive drinking will damage the liver, stomach, spleen and other organs, even lead to neurological diseases, which has become a problem of great concern. Scientists have pointed out that ethanol is highly likely to affect gene expression in the body, which provides a basis for studying both the pathological and the addictive characteristics of alcohol. As a special model organism, C. elegans has relatively simple and clear physiological structure (especially its neural structure), and a large number of confirmed homologous genes that associated with human diseases. Therefore, it is used as a biological model to study the molecular targets and related neural pathways of disease occurrence. Recently, Mark G. et al. published a transcriptional analysis of ethanol exposure in C. elegans, illustrating the mechanism of action.
The transcriptional responses of C. elegans exposed to ethanol were analyzed in this paper. In order to achieve the experimental purpose, the experiment of time process was carried out. C. elegans were respectively exposed to 0 mM or 400 mM ethanol for 0, 30, 60, 120, or 480 mins. The RNA was isolated and the transcriptional response to ethanol was analyzed by microarrays. It was found that different durations of ethanol exposure triggered different transcription results (Fig.1). During the 30min ethanol exposure, an increase in the expression of mod-1 induced by ethanol could be observed. This gene can encode and express serotonin, which is a mechanism for animals to regulate behavior according to external stimuli is also related to human alcohol use disorder. However, at the time point of 480 min, it was observed that ethanol exposure induced an overall reduction in serotonin signaling. The genes involved in alcohol metabolism were highly expressed at 60 min. While the long-term ethanol exposure has a much greater impact on transcriptome. Several genes known to be involved in the physiological response of ethanol, including direct ethanol molecular targets, were regulated at 8 h of ethanol exposure. At the same time, too long exposure to ethanol led to the regulation of genes related to cilia function. In human body the Alcohol Induced Ciliary Dysfunction (AICD) leads to severe lung disease in many long-term alcoholics. During the food deprivation experiment, the authors also found that C. elegans can use ethanol as the source of calories.
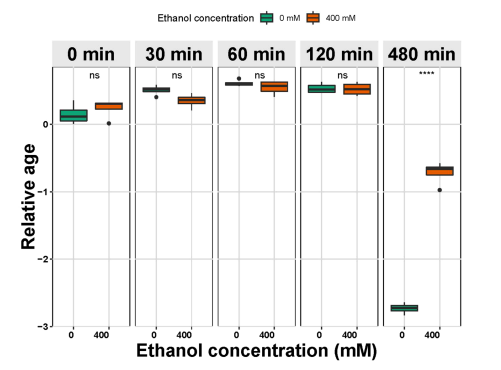
Figure 1. Effect of ethanol exposure on transcriptional time, presented as relative age.
Through the above experimental results, we can find that ethanol exposure can affect the expression of genes related to transcriptional regulatory pathways in vivo. The construction of this nematode model can help us study the regulation of genes in long-term alcoholics by ethanol exposure resulting serious health effects. The current related research needs to be further developed.
Reference: Transcriptional analysis of the response of C. elegans to ethanol exposure[J]. Scientific Reports, 2021, 11(1).
doi: 10.1038/s41598-021-90282-8
Keywords:C. elegans, ethanol exposure, transcriptome, transcriptional analysis, mod-1, serotonin, cilia function.



