Role of ubiquitination in the aging process of Caenorhabditis elegans
2021-08-06 15:28
Biologically, aging is a spontaneous and inevitable process of life over time. It is a complex natural phenomenon, characterized by structural degeneration and functional decline, with the decrease of adaptability and resistance of the body. We can simply think that aging is caused by a loss of cellular integrity. In view of the important role of ubiquitin modifications in cell function, many scientists have linked ubiquitination with aging to study the aging phenomenon of the body from a more subtle level.
Recently, a study on the relationship between ubiquitination and aging using C. elegans has been published in Nature. The paper assesses the link between ubiquitination and aging by quantifying whole-proteome ubiquitin signatures in C. elegans. The authors demonstrate from multiple perspectives that the overall deubiquitination across tissues during aging will damage the degradation of targeted proteasomes of lifespan regulators.
The paper has shown that ageing causes a global loss of ubiquitination that is triggered by increased deubiquitinase activity. Authors assessed age-matched long-lived genetic models of dietary restriction (eat-2(ad1116)) and reduced insulin/IGF-1 signalling (daf-2(e1370)) (Fig.1 A). C. elegans undergoes a widespread proteome remodelling during ageing. In many instances, differences in ubiquitination levels could not be simply ascribed to a similar change in the protein amounts. Experimental results show that the dietary restriction rescued the levels of CSN-6, a component of the COP9 signallosome that removes ubiquitin-like proteins from cullin E3 complexes, inhibiting their activity and the subsequent ubiquitination of proteasomal targets. In addition to CSN-6, reduced insulin/IGF-1 signalling prevented the upregulation of most of the age-dysregulated DUBs (Fig.1 B). Knockdown of distinct age-dysregulated DUBs ameliorated loss of ubiquitination during aging, including csn-6(which encodes the worm orthologue of human COPS6), H34C03.2 (USP4), F07A11.4 (USP19), math-33 (USP7), usp-5 (USP5), usp-48 (USP48) and otub-3 (OTUD6A). The researchers also found that using DUB inhibitors in old worms was sufficient to rescue low ubiquitination levels and extend lifespan (Fig.1 C).
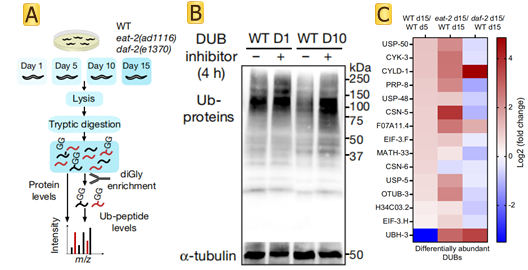
Fig1 DUBs diminish ubiquitination with age
Ubiquitination can tag proteins for recognition by the proteasome, but the researchers found that 192 proteins in the experimental process of aging at least one less lysine locus of ubiquitin. This is likely because the related protease activity caused by the defects of body, so there is a fundamental question is whether the defect of targeted degradation influence longevity. By integrating data from worms with a defective proteasome, the researchers found that 10 proteasome-modulated proteins became more abundant with age, and at least one of their lysine sites was less ubiquitinated (Fig.2 A). Lowering the levels of age-dysregulated proteasome targets prolongs longevity, whereas preventing their degradation shortens lifespan. These targets were IFB-2, EPS-8, RPL-4, M01G12.9, C46C2.2, F54D1.6, DDI-1, LEC-1, HSP-43 and USP-5. The author focuses on the IFB-2 intermediate filament and the EPS-8 modulator of RAC signalling. For more thorough research authors customize the ubiquitin-less IFB-2 and EPS-8 mutant worms made by SunyBiotech using CRISPR-Cas9 methodology. Intrigued by the robust effects of IFB-2 and EPS-8 on lifespan, western blot was used to confirm that their protein levels increased with age (Fig.2 B,C). The bioinformatic analysis of tissues such as the germ line, muscle, intestine, epidermis or neurons reveals that ubiquitination loss occurs throughout the organism (Fig.2 D). The amounts of the ubiquitously expressed EPS-8 protein were increased in different somatic tissues with age (Fig.2 E), which correlates with global deubiquitination across tissues. The researchers blocked the ubiquitination of endogenous IFB-2 and EPS-8 by generating lysine (K) to arginine (R) mutants. An important finding in the experiment was that the K255R/K341R double mutation increased IFB-2 protein levels in young adult worms and shortened lifespan (Fig.2 F-H). Notably, knockdown of eps-8 after development was sufficient to rescue the short-lived phenotype of EPS-8 (K524R/K583R/K621R) mutants (Fig.2 I). These results suggest that the age-related deubiquitination and subsequent impaired degradation of IFB-2 and EPS-8 determine lifespan.
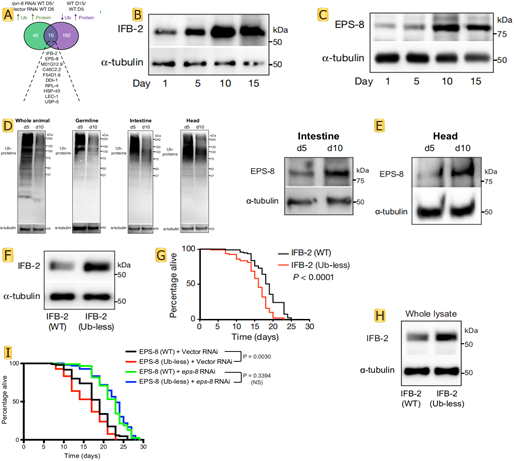
Fig.2 Proteasome targets determine lifespan
Further exploration of the functions of IFB-2 and EPS-8 revealed that increased levels of IFB-2 promote the loss of intestinal integrity and bacterial colonization, upregulation of EPS-8 hyperactivates RAC in muscle and neurons (Fig.3 A-C), and leads to alterations in the actin cytoskeleton and protein kinase JNK (Fig.3 D-F). In summary, age-related changes in targeted degradation of structural and regulatory proteins across tissues determine longevity.
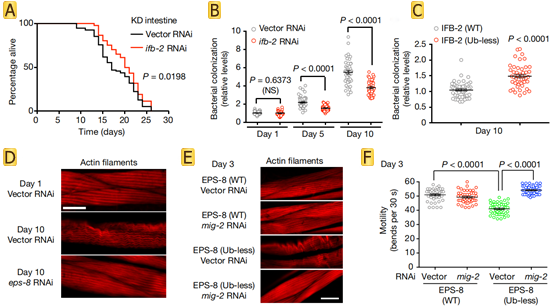
Fig.3 Increased IFB-2 levels induce age-related intestinal alterations and increased EPS-8 levels shorten lifespan through RAC hyperactivation
Reference: Koyuncu S, Loureiro R, Lee H J, et al. Rewiring of the ubiquitinated proteome determines ageing in C. elegans[J]. Nature, 2021: 1-6.
Doi: 10.1038/s41586-021-03781-z





