Decoding Sex-Specific Gene Networks in C. elegans: A New Atlas of Dimorphic Gene Expression
2024-09-26 10:32
Keywords: Sex-specific gene expression, C. elegans, developmental biology, INS-39, gene networks
Contribution of SunyBiotech
SunyBiotech was instrumental in creating the essential genetic tools that enabled the study of sex-specific gene expression in C. elegans.
The specific alleles generated by SunyBiotech include:
•ins-39(syb4915[ins-39::SL2::GFP::H2B])
•srj-49(syb4956[srj-49::SL2::GFP::H2B])
•ces-2(syb4992[ces-2::SL2::GFP::H2B])
Introduction
The nematode Caenorhabditis elegans (C. elegans) is an invaluable model for studying sex-specific gene expression and developmental biology. As a hermaphoditic species with males, it provides a unique opportunity to study the dimorphic gene networks that influence sex-specific traits. Despite significant advances, there has been a lack of comprehensive datasets analyzing gene expression in both sexes throughout various developmental stages. This gap hampers the ability to draw conclusions about how sexual dimorphism influences development.
Recent studies have begun to illuminate the molecular underpinnings of sex-specific development, but the need for a transcriptomic atlas that comprehensively maps gene expression across both sexes and multiple developmental stages remained unmet. This study seeks to fill that gap by providing a detailed gene expression atlas that tracks developmental changes from early larval stages to adulthood in both male and hermaphroditic C. elegans.
The Study: Objectives and Key Findings
Objectives
The study aimed to develop a comprehensive developmental gene expression atlas for both sexes of C. elegans to map the dimorphic regulatory networks across various life stages. The focus was particularly on identifying and understanding the role of differentially expressed genes (DEGs) that exhibit sex-specific regulation. One of the key genes examined was ins-39, which was analyzed in the context of its expression patterns and impact on survival, stress response, and sexual differentiation.
Key Findings
Sexually Dimorphic Gene Expression Throughout Development
The gene expression data covered five key developmental stages: L1, L2, L3, L4, and young adults (YA). Across these stages, 14,185 genes were profiled, of which 33% showed sex-specific expression at one or more stages. At the L1 stage, 519 genes were differentially expressed between males and hermaphrodites, and this number gradually increased, peaking in young adults with 3,564 differentially expressed genes (DEGs) (Figure 1). This pattern highlights the progressive divergence in gene regulation as sexual maturity approaches, with males showing a larger bias in gene expression throughout development. The researchers also found that male-biased genes outnumbered hermaphrodite-biased genes in most stages, suggesting a greater diversity in male-specific gene regulatory mechanisms.
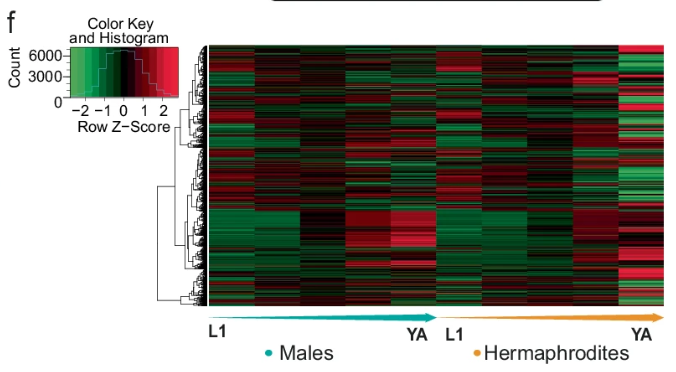
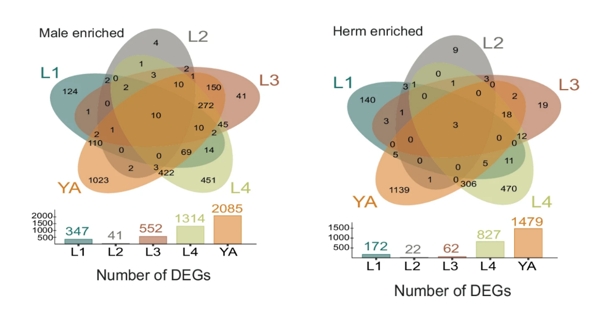
Figure:1: A remarkable number of genes were found to exhibit sexually dimorphic expression patterns at various stages of development.
Distinct Patterns of ins-39 Expression in Males and Hermaphrodites
One of the standout discoveries was the sexually dimorphic expression of the ins-39 gene, an insulin-like peptide, which exhibited consistently higher expression in males across all developmental stages. The quantitative comparison of ins-39 expression between sexes showed a dramatic difference, with male showing higher expression levels than hermaphrodites (Figure 2). The expression of ins-39 in neurons was mapped through confocal imaging, revealing that in males, the gene is expressed in a broader array of sensory neurons (AFD, ASJ, ASK, and others), while in hermaphrodites, it is restricted to fewer neurons (Figure 2).
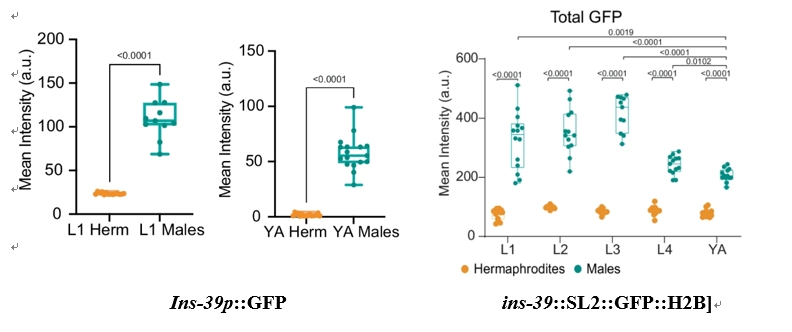
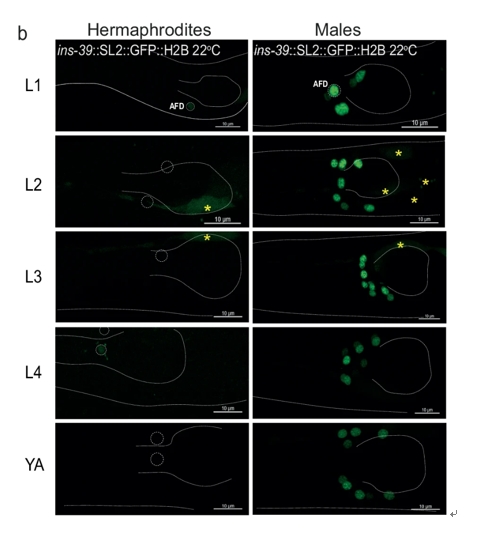
Figure 2: Distinct Patterns of ins-39 Expression in Males and Hermaphrodites
Role of ins-39 in Stress Responses and Survival
The role of ins-39 was further examined in the context of survival under stress. In L1 stage survival assays, wild-type males showed lower survival rates compared to hermaphrodites, but when ins-39 was knocked out, both sexes exhibited significantly enhanced survival during the L1 arrest phase. Figure 3 illustrated this phenomenon, where males particularly benefited from the absence of ins-39, surviving longer under L1-arrest conditions than wild-type males, reinforcing the gene’s role as a negative regulator of L1 survival.
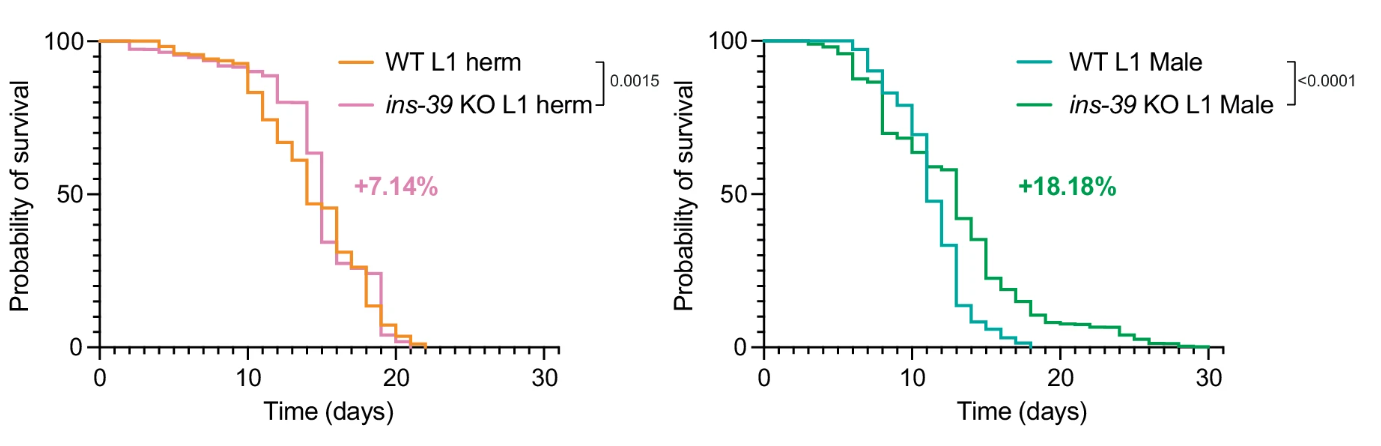
Figure 3: Males particularly benefited from the absence of ins-39, surviving longer under L1-arrest conditions than wild-type males
The gene’s role in stress response was also tested using oxidative stress assays. The findings showed that ins-39 influenced hermaphrodites' survival under oxidative stress, as seen in the increased survival of ins-39 mutant hermaphrodites when exposed to tert-butyl hydroperoxide (Figure 4). However, males exhibited no such dependence on ins-39 for stress response, reinforcing the idea that the gene plays a sex-specific role, primarily affecting hermaphrodites in stressful conditions.
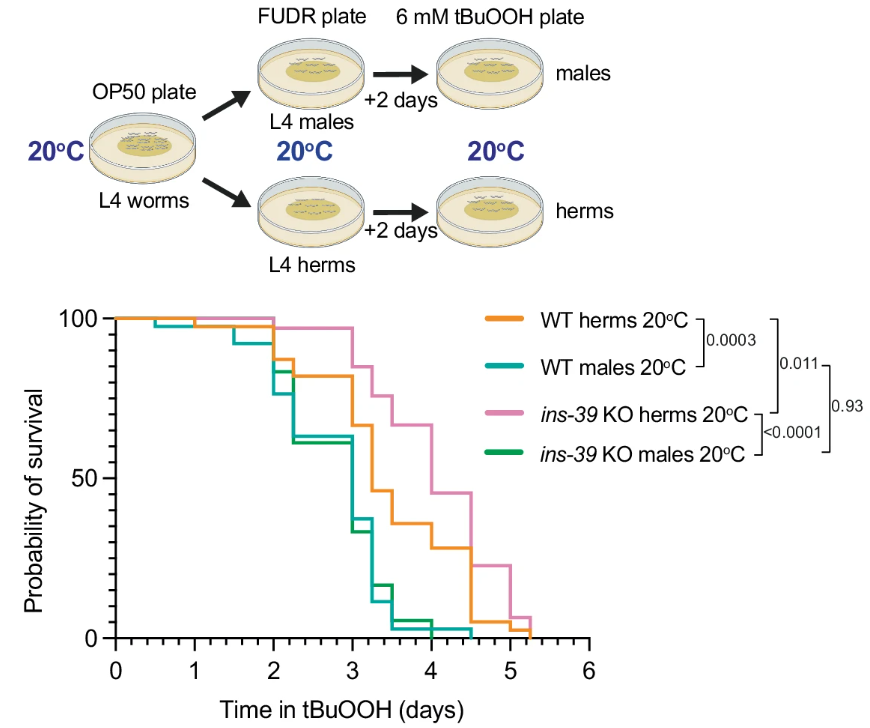
Figure 4: ins-39-dependent stress responses were evident in hermaphrodites but absent in males
Dimorphic Neuropeptide Gene Expression
Beyond ins-39, the study revealed extensive dimorphic regulation of neuropeptide genes. These genes, which are essential for neuronal signaling, showed male-biased expression patterns, especially at later stages of development. Of the 37 neuropeptide genes with dimorphic expression, only a handful showed significant differences before the L4 stage, indicating that most neuropeptide-driven differences emerge after sexual maturation (Figure 5). This delayed onset of neuropeptide dimorphism points to a critical role for these genes in shaping sex-specific behaviors and neural circuits later in development.
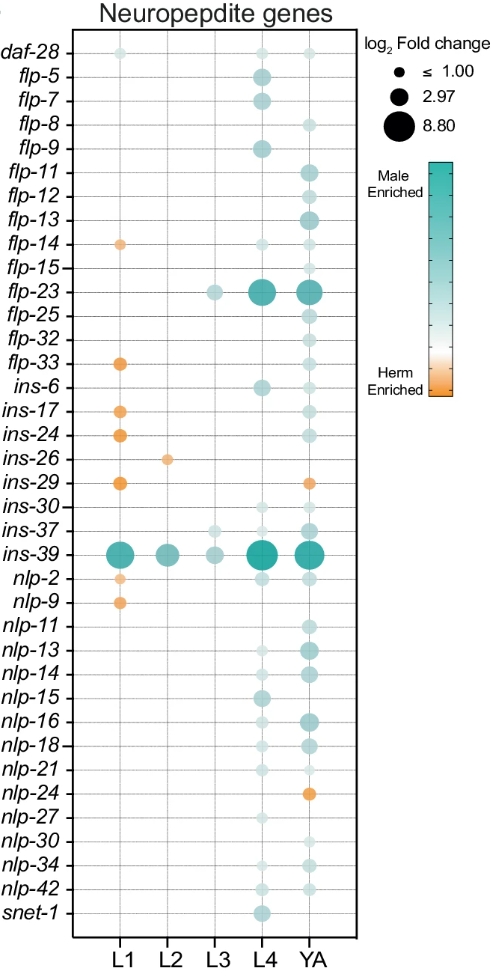
Figure 5 The most neuropeptide-driven differences emerge after sexual maturation
Sex-Specific Transcriptional Regulation
The study also investigated the broader regulatory networks driving dimorphic gene expression. Transcription factors like TRA-1 and MAB-3 were identified as key regulators of sex-specific gene expression. TRA-1, known for its role in repressing male-specific traits, was found to restrict ins-39 expression in hermaphrodites, while MAB-3 acted as a promoter of ins-39 in males (Figure 6). The findings suggest a complex regulatory system where sex-specific transcription factors either activate or repress critical genes, depending on the organism’s genetic sex.
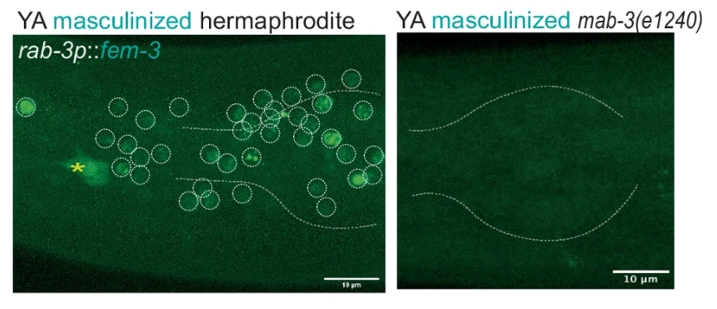
Figure 6:MAB-3 acted as a promoter of ins-39 in males
Conclusion
This study offers a robust, stage-by-stage map of sex-specific gene expression in C. elegans, highlighting key differences between males and hermaphrodites at each developmental stage. By focusing on genes like ins-39, which play pivotal roles in stress response and survival, the research provides insights into the molecular mechanisms that govern sexual dimorphism (Figure 7). The findings not only expand our understanding of gene regulation in C. elegans but also lay the groundwork for exploring similar processes in other organisms.
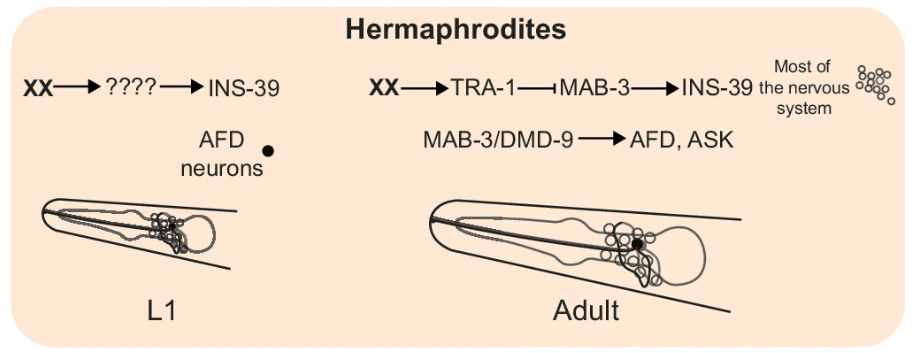
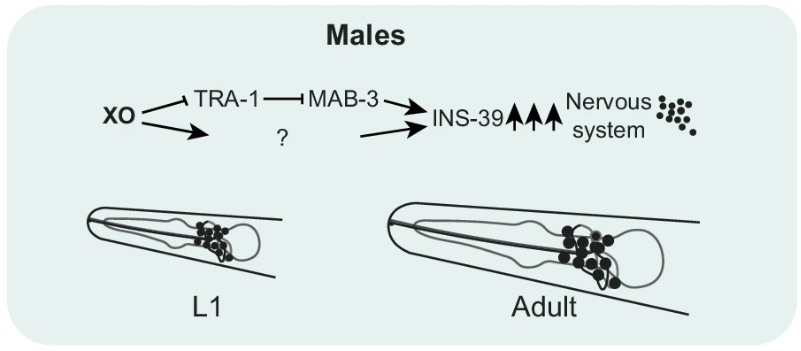
Figure 7: A model depicting the molecular elements mediating INS-39 expression in hermaphrodites and males at L1 and YA stages
References
Haque, R., Kurien, S. P., Setty, H., et al. (2024). Sex-specific developmental gene expression atlas unveils dimorphic gene networks in C. elegans. Nature Communications, 15:4273. https://doi.org/10.1038/s41467-024-48369-z.












