Disease-associated Mutations in CHIP: A Key Regulator of Mitophagy
2024-12-03 11:43
Keywords: mitophagy, CHIP mutations, STUB1, neurodegeneration, SCA48, ataxia
Introduction
A study in Neurobiology of Disease has shed light on the role of C-terminus of HSP70 interacting protein (CHIP) in the regulation of mitophagy, a process critical for maintaining cellular health by removing damaged mitochondria. CHIP, an E3 ubiquitin ligase and HSP70 cochaperone, has been linked to neurodegenerative diseases such as spinocerebellar ataxia autosomal dominant type 48 (SCA48) and autosomal recessive type 16 (SCAR16). CHIP deficiency leads to increased levels of the proteins PTEN-induced kinase 1 (PINK1) and Parkin, which are both key regulatory proteins in the PINK1/Parkin mitophagy pathway. The research provides insights into how mutations in CHIP can impair its ability to negatively regulate mitophagy, potentially contributing to disease progression.
Contribution of SunyBiotech:
SunyBiotech constructed two disease-associated Caenorhabditis elegans mutant strains for studying the ability of CHIP mutations to regulate mitophagy in vivo:
PHX5417:chn-1(syb5417)
PHX5418:chn-1(syb5418)
1. CHIP is a negative regulator of the PINK1/Parkin mitophagy pathway
After treatment with CCCP, overexpression of CHIP in vitro decreased PINK1 protein levels, the percentage of cells undergoing mitophagy, and the percentage of cells with mitochondrial autophagy compared to controls(Fig. 1A, 1B and 1C). Similarly, knockdown of endogenous CHIP increased the percentage of cells with Parkin punctalocated in mitochondria, and the percentage of cells with mitochondrial autophagy(Fig. 1D, 1E and 1F). These results showed that under normal conditions, CHIP promoted the degradation of PINK1, impaired Parkin translocation to the mitochondria, and suppressed mitophagy in response to mitochondrial stress.
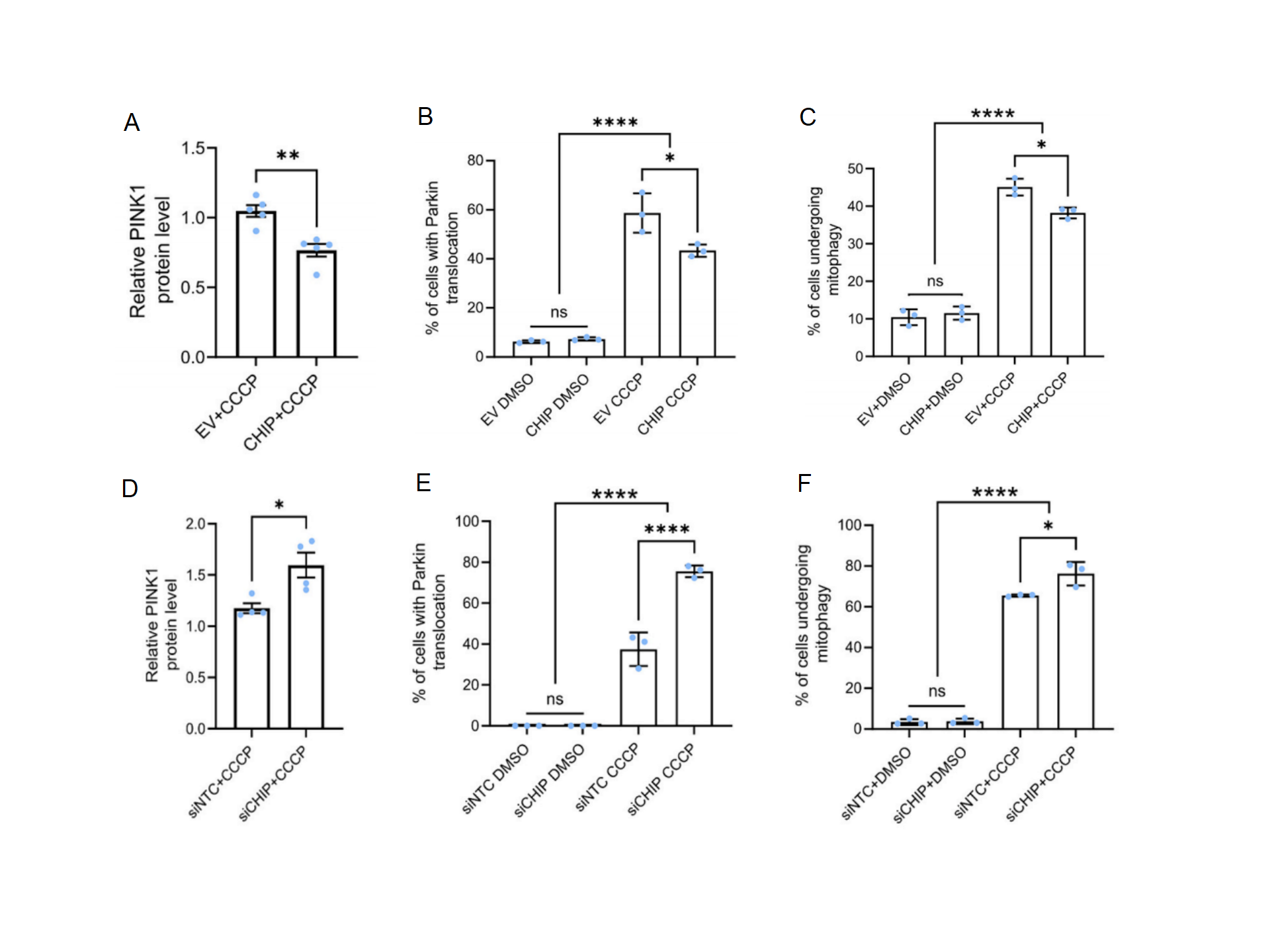
Figure 1. CHIP is a negative regulator of the PINK1/Parkin mitophagy pathway
2. Loss of CHIP increased neuronal mitophagy in C. elegans in a PINK1/Parkin-dependent manner
The study revealed that loss of CHIP function, due to disease-associated mutations, enhances neuronal mitophagy in a PINK1 and Parkin-dependent manner in C. elegans. In CHIP-deficient chn-1(by155) nematodes, there was an increase in mitochondrial co-localization with phagosomes in neurons (Fig. 2A), and elevated levels of mitochondrial autophagy (Fig. 2B). Pink-1(tm1779) and pdr-1(gk488) neurons with deletion of PINK1 and Parkin homologues showed reduced autophagy. However, the GFP/DsRed intensity ratio was increased in the progeny of crosses with chn-1 (by155), suggesting that the increase in neuronal autophagy observed in CHIP-deficient nematodes is dependent on PINK1 and Parkin (Figure 2B). These findings suggest that mutations in CHIP can dysregulate mitophagy both in vitro and in vivo, potentially leading to the mitochondrial dysfunction observed in neurodegenerative diseases.
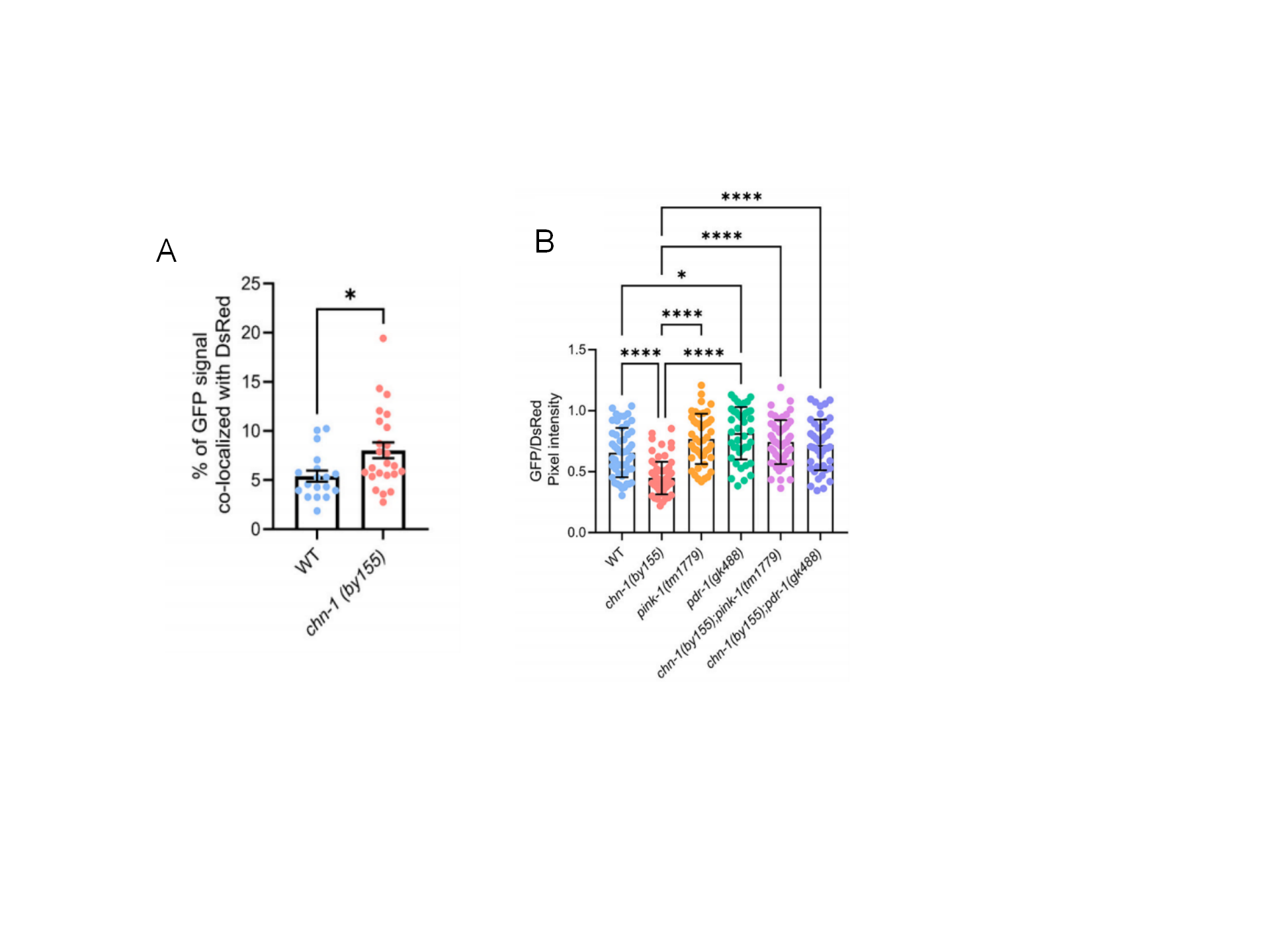
Figure 2. Loss of CHIP increased neuronal mitophagy in C. elegans
3. Impact of disease-associated mutations and neurodegenerative diseases
It has been reported that many CHIP mutations linked to diseases impair CHIP functionality. This article investigated the effects of two related mutations, I53T and L275Dfs*16, on CHIP's regulation of mitophagy. Neither of them resulted in a decrease in PINK1 protein levels(Fig. 3A). Interestingly, overexpression of WT CHIP significantly reduced the percentage of mitochondrial autophagic cells after CCCP treatment, whereas I53T or L275Dfs*16 did not (Fig. 3B). CRISPRCas9 was used to construct mutant nematodes CHN-1 I32T (chn-1 (syb5417)) and CHN-1 L233Yfs*2 (chn-1 (syb5418)) to investigate whether the two disease-associated mutations in CHIP affect mitochondrial autophagy in vivo. The results showed that all CHIP-associated mutations increased neuronal autophagy(Fig. 3C). It is possible that there are differences in basal mitophagy, suggesting that some basal PINK1/Parkin-mediated mitophagy occurs in C. elegans.
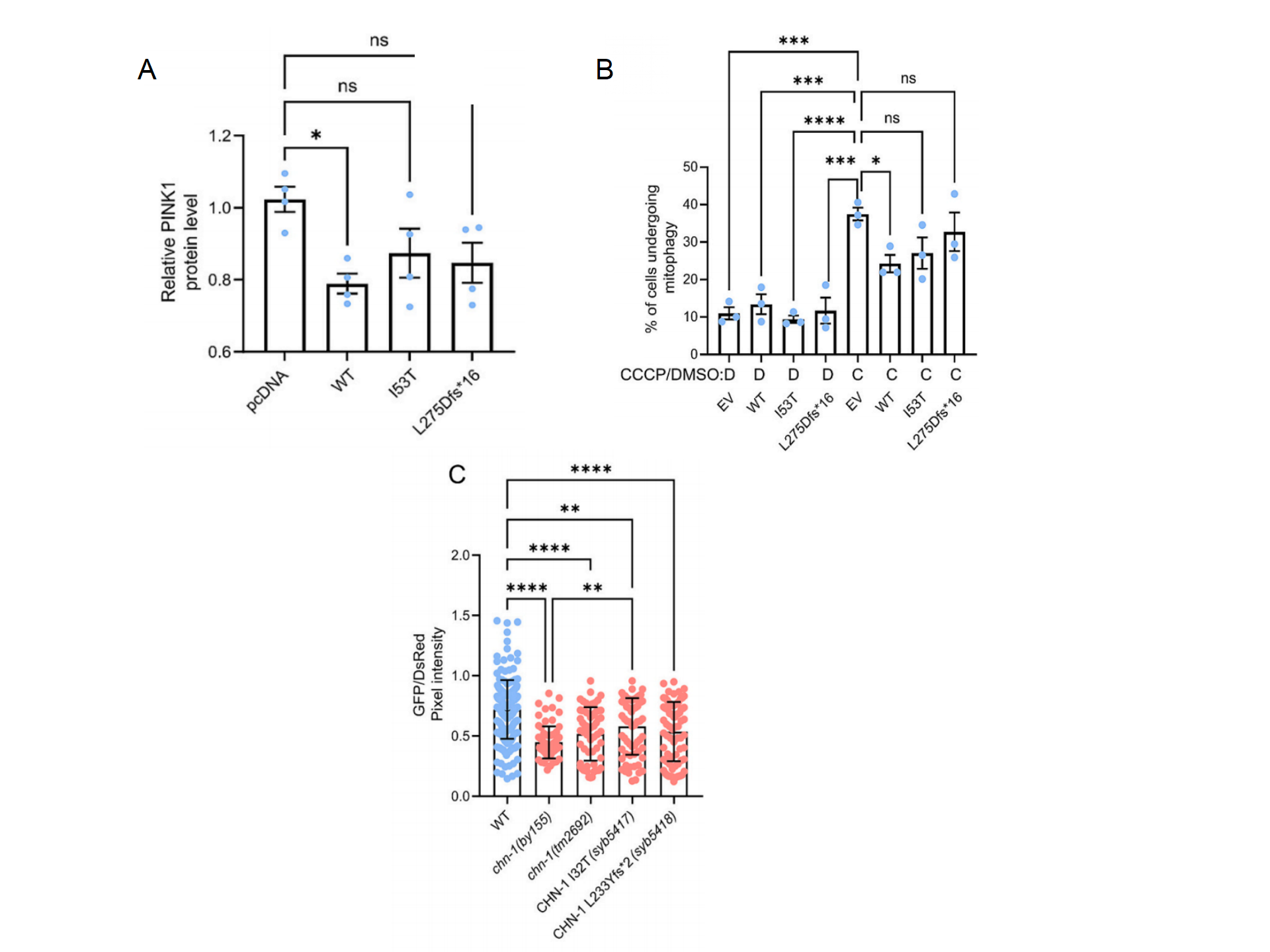
Figure 3. Impact of disease-associated mutations and neurodegenerative diseases
Conclusion
The study by Rebecca Earnshaw et al. provides a deeper understanding of the role of CHIP in mitophagy and its implications in neurodegenerative diseases. The research highlights the importance of CHIP in maintaining mitochondrial health and suggests that dysregulation of mitophagy due to CHIP mutations could be a contributing factor in the development of neurodegenerative diseases. This discovery could have significant implications for the development of therapeutic strategies targeting CHIP and mitophagy in diseases like SCA48 and SCAR16.
References
Earnshaw R, et al. Disease-associated mutations in C-terminus of HSP70 interacting protein (CHIP) impair its ability to negatively regulate mitophagy. Neurobiol Dis. 2024 Oct 1:200:106625.





