Cracking the Genetic Code of Behavior: How Variants in C. elegans Shape Foraging
2025-02-10 10:13
Keywords
Genetic Variation, Foraging Behavior, Neuroendocrine Signaling, daf-7, gap-2
Contribution of SunyBiotech
This cutting-edge research leveraged advanced genetic tools provided by SunyBiotech to dissect the molecular pathways linking genetic variation to behavior. SunyBiotech’s expertise enabled precise genome editing and analysis, revealing how natural variants in the gap-2 gene regulate neuroendocrine signaling and behavioral adaptation.
The specific strains generated by SunyBiotech:
ZD2683 ksIs2;gap-2(syb4046)
PHX4046 gap-2(syb4046)
PHX5109 gap-2(syb5109)
PHX5128 gap-2(syb5128)
PHX6873 gap-2(syb6873)
PHX4684 gap-2(syb4684)
PHX4587 gap-2(syb4587)
PHX5834 gap-2(syb4587 syb5834)
Introduction
Understanding the genetic basis of behavior is one of biology’s great challenges. In Caenorhabditis elegans, a nematode with diverse feeding behaviors, the neuroendocrine ligand daf-7 plays a central role in translating environmental cues into foraging decisions. This study reveals how natural variants in the gap-2 gene influence daf-7 expression in specific neurons, shaping the balance between exploratory roaming and stationary dwelling during foraging.
The Study: Objectives and Key Findings
Objectives
· Identify natural genetic variants affecting daf-7 expression in C. elegans.
· Investigate how these variants influence foraging behavior.
· Establish the molecular mechanisms linking genetic variation, neuroendocrine signaling, and behavior.
Key Findings
1. Genetic Variation in daf-7 Expression
The study began by surveying wild strains of C. elegans to identify differences in daf-7 expression within ASJ neurons, known to mediate foraging behavior. A striking difference emerged between the N2 and MY18 strains, with MY18 exhibiting significantly higher daf-7 expression. Genome-wide linkage analysis revealed a quantitative trait locus (QTL) on chromosome X, which explained 11.48% of the variation in daf-7 expression (Figure. 1). This finding prompted an investigation into the specific genetic variants within the QTL affecting daf-7 expression.
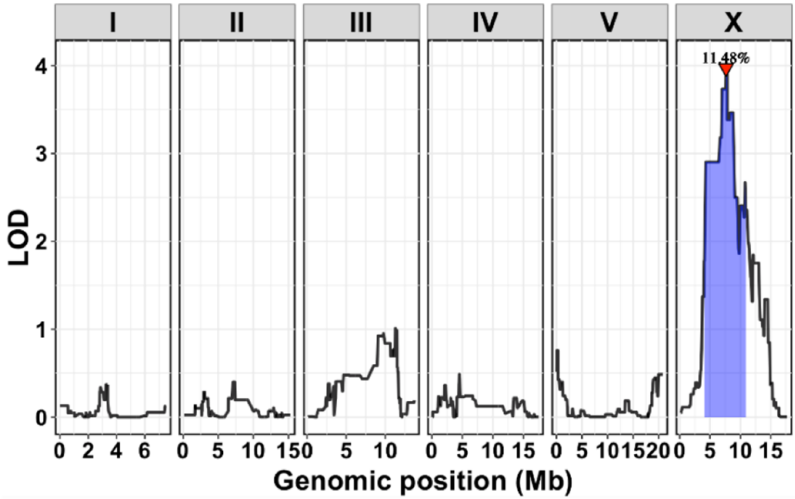
Figure 1: Linkage Mapping of daf-7p::GFP Expression in ASJ Neurons
2. Role of gap-2 Variants
Within the QTL, researchers identified gap-2, a gene encoding a Ras GTPase-activating protein, as a critical modulator of daf-7 expression. Specific variants, such as 64T and 11L, were found to increase daf-7 expression. Strains carrying the 64T variant (Figure. 2a and b) exhibited upregulated daf-7 expression in ASJ neurons, while engineering a reciprocal mutation (64S) reduced this expression (Figure. 2c). It has established a connection between gap-2 variants and daf-7 expression, the study explored their impact on behavior.
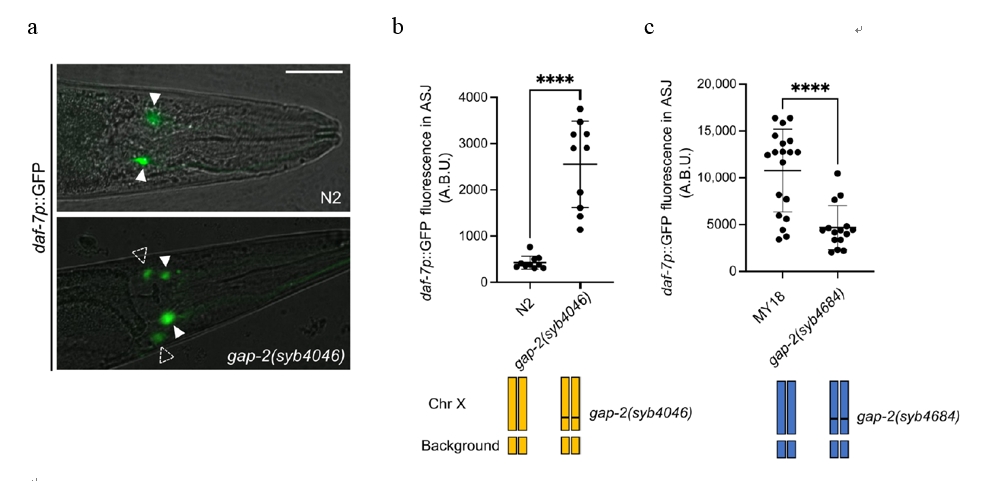
Figure 2: Daf-7p::GFP Expression in N2, gap-2(syb4046), and gap(sy4684) Strains
3. gap-2 Variants Drive Roaming Behavior in Distinct Genetic Backgrounds
The roaming-promoting effects of gap-2 variants were context-dependent. In the N2 background, the 64T variant increased roaming, whereas introducing the 64S variant into the MY18 background reduced roaming relative to its native 64T variant. Strains carrying the 64T variant in the N2 background spent more time roaming, while MY18 strains engineered to carry the 64S variant showed reduced roaming compared to native MY18 strains (Figure. 3).
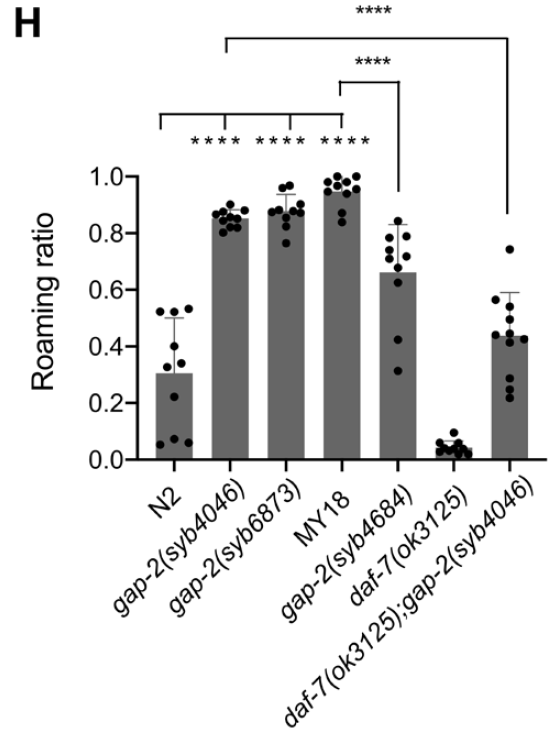
Figure 3: Effect of gap-2 Variants on Roaming Behavior Across Genetic Backgrounds
4. Cell-Nonautonomous Modulation of Neuroendocrine Signaling
To understand how these behavioral changes occurred, the researchers examined the cellular mechanisms driving gap-2 function. The gap-2 variants acted in neurons outside the ASJ pair, particularly the ADE sensory neurons, to influence daf-7 expression. Expressing the 64T variant in ADE neurons alone was sufficient to increase daf-7 expression in ASJ neurons, suggesting a cell-nonautonomous regulatory mechanism (Figure. 4). These findings raised questions about the evolutionary and ecological relevance of these genetic variants.
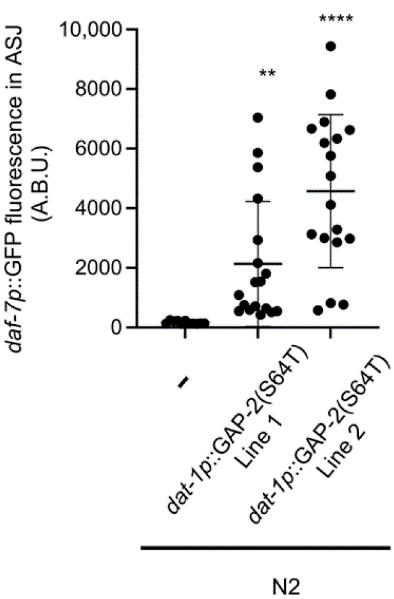
Figure 4: Maximum daf-7p::GFP Fluorescence in ASJ Neurons of ADE Neuronal GAP-2(64T) Transgenics
5. Ecological and Evolutionary Context
The 64T and 11L variants were enriched in strains isolated from compost environments, suggesting an adaptive role in enabling flexible foraging in dynamic microbial landscapes. Statistical analysis revealed a significant association between the 64T variant and strains from compost substrates compared to other environments (Figure. 5).
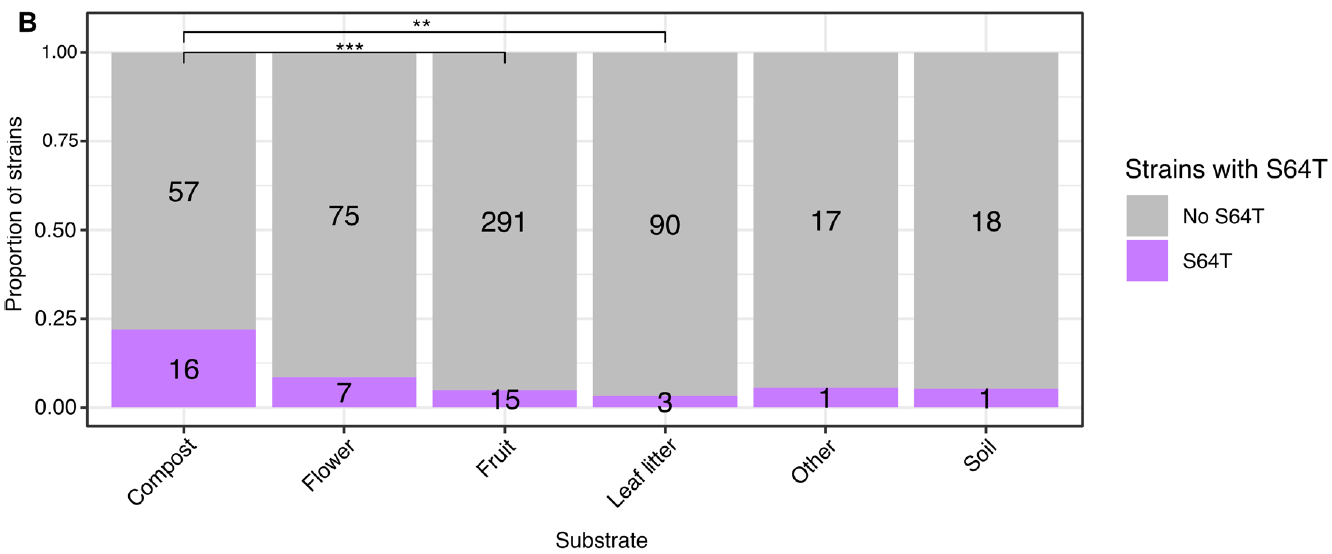
Figure 5: Substrate Enrichment of S64T in Wild-Type Strains
Conclusion
This study illuminates how natural variants in gap-2 shape the expression of daf-7, a neuroendocrine regulator, to influence foraging behavior in C. elegans. By linking genetic variation to behavioral adaptability, these findings deepen our understanding of host-microbe interactions and evolutionary ecology.
References
Lee, H., Boor, S. A., Hilbert, Z. A., et al. (2024). Genetic variants that modify neuroendocrine gene expression and foraging behavior of C. elegans. Science Advances, 10, eadk9481. DOI:10.1126/sciadv.adk9481.







