Gene knockout
Gene knockout is an important genome editing technology. When the gene is deleted, observation of the phenotype of knockout can reveal the role played by the gene in the organism, the role in life activities and further reveal its role in human diseases. Up to now, SunyBiotech can efficiently knock out 43,000 bp of nematode genome through CRISPR/Cas9 system.
Gene knockout has incomparable high efficiency compared with RNA interference (RNAi). It not only overcomes the off-target effect in the process of RNAi but also can generate stable genetic mutant because the genome sequence is permanent changed by gene knockout. Furthermore, mutated strains of specific genes obtained through gene knockout can also hybridize with mutant nematodes of other genes to obtain strains with double mutations or even multiple mutations. Therefore, gene knockout can be used in the research on the interaction of multiple proteins and greatly advances the research and development of related functional genes.
Our advantage
The principle of CRISPR-Cas9 system is simple. The key influencing factors include Cas9 protein, sgRNA and template DNA, each one of which have impact on accuracy and efficiency of gene editing Especially, how to accurately delete large DNA fragments has always been a great challenge in gene editing.
Sunybiotech has made many improvements in the application of CRISPR/Cas9 system in order to obtain the required mutants more efficiently and precisely. At present, we have completed 206 gene knockout orders with a success rate of 100%, 18 of which are large fragment knockout (5000bp-43,000bp), The details are shown in Table 1.
Table 1 Precise sequence deletion service of SunyBiotech
KO segment length (bp) | Cases of service | Success rate |
≤5,000 | 188 | 100% |
5,000-12,000 | 11 | 100% |
12,000-20,000 | 3 | 100% |
30,000-43,000 | 4 | 100% |
It’s worthy mentioning that SunyBiotech has successfully generate the ptp-3 null mutant (more than 43kb) using CRISPR/Cas9 gene editing system (Figure. 1).
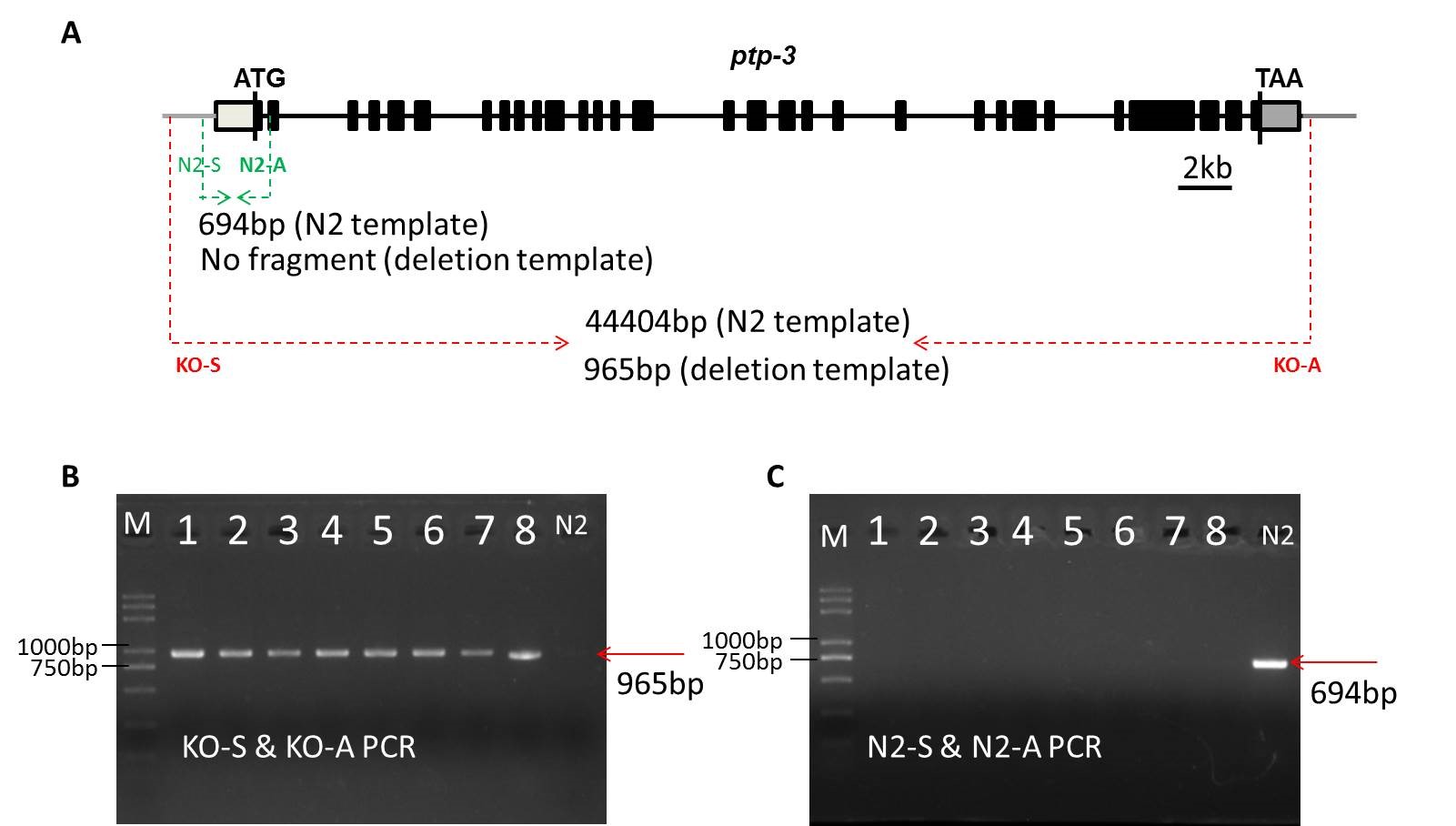
Figure. 1 Generation of the ptp-3 null mutant
A: The schematic diagram of the ptp-3 gene. The black blocks indicate exons and black lines indicate introns, and the annealing site of genotyping primers have been marked.
B, C: The gel figures of homozygotes genotyping by using the outside (KO-S & KO-A) primer pair (B) and the inside (N2-S & N2-A) primer pair.
Using a pair of outside primers and a pair of inside primers for genotyping, we determined that the screened knockout nematode is homozygous and confirmed the precise ptp-3 null mutant by sequencing. In order to make sure that the deleted 43kb DNA does not form extrachromosomal arraies, we designed a pair of primers that anneals to the deleted sequence to determine that the homozygous knockout strain does not contain deleted DNA sequence.



