The Secrets of Mitochondrial Inheritance: How Cells Selectively Destroy Paternal Mitochondria
2024-10-22 10:35
Keywords: ALLO-1, IKKE-1, mitochondrial inheritance, autophagosome, paternal mitochondrial degradation
Contribution of SUNY Biotech:
SunyBiotech was instrumental in creating the essential genetic tools that enabled the study of selective autophagy in C. elegans.
The specific alleles generated by SunyBiotech include:
ikke-1(syb2844)
unc-51(syb2940)
Introduction:
Mitochondrial inheritance is a cornerstone of cellular biology, with the maternal line exclusively passing on mitochondrial DNA (mtDNA) to offspring. However, the mechanisms that ensure the elimination of paternal mitochondria have long intrigued scientists. Recent research on C elegans offers a breakthrough in understanding how selective autophagy clears paternal mitochondria post-fertilization. By investigating the roles of two key proteins— ALLO-1, a cytosolic autophagy adaptor, and IKKE-1, a kinase, coordinate the selective degradation of paternal mitochondria—this study reveals a sophisticated feedback mechanism that triggers autophagosome formation, facilitating the degradation of paternal organelles. The findings not only illuminate fundamental processes in developmental biology but also hold promise for therapeutic applications in mitochondrial diseases.
The Study: Objectives and Key Findings
Objectives:
The study aimed to explore how ALLO-1 and IKKE-1 in C. elegans embryos. The researchers sought to clarify how these proteins interact to regulate the initiation of autophagy, ensuring that only paternal organelles are targeted for degradation, while maternally inherited mitochondria remain intact.
Key Findings:
ALLO-1 Isoforms and Their Targeting of Paternal Organelles
One of the key findings of the study was that ALLO-1 exists in two isoforms, ALLO-1a and ALLO-1b, which have distinct roles in selective autophagy. ALLO-1b was found to target paternal mitochondria, while ALLO-1a primarily localized around membranous organelles (MOs) from sperm. This suggests that different components of the paternal cellular machinery are recognized and degraded through separate pathways. The live imaging data in Figure 1 vividly demonstrate the preferential localization of ALLO-1b around paternal mitochondria, highlighted by the green fluorescence signal, while ALLO-1a, as shown in Figure 1, primarily associates with MOs.
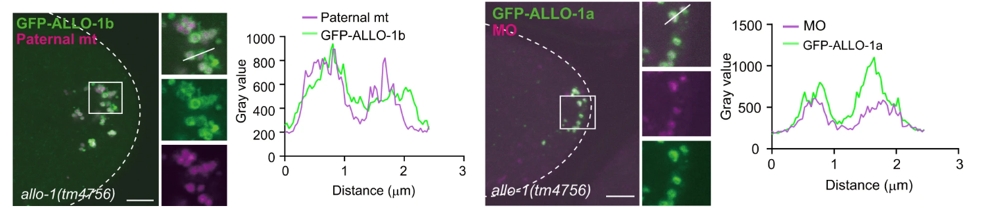
Figure 1: The preferential localization of ALLO-1b around paternal mitochondria, while ALLO-1a primarily associates with Mos
IKKE-1’s Role in Supporting ALLO-1b Accumulation
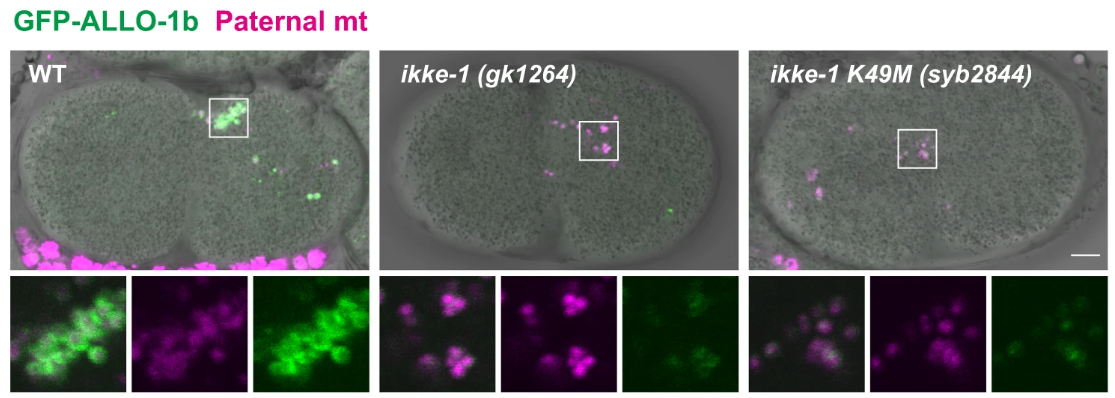
As paternal mitochondria are recognized by ALLO-1b, a critical observation was made regarding the role of the kinase IKKE-1 in this process. While ALLO-1b could still recognize mitochondria in ikke-1 mutant embryos, it failed to accumulate around the organelles in sufficient quantities to promote autophagosome formation. This finding highlighted the importance of IKKE-1 in enabling the progression from recognition to degradation. In wild-type embryos, GFP-tagged ALLO-1b shows robust accumulation around the paternal mitochondria, whereas in ikke-1 mutants, the intensity of ALLO-1b is significantly reduced (Figure 2a). This impaired accumulation was also captured through time-lapse imaging in Figure 2b, which shows delayed and incomplete ALLO-1b localization in ikke-1 mutants. This data points to IKKE-1’s essential role in sustaining the autophagic response necessary for mitochondrial degradation.
a
b
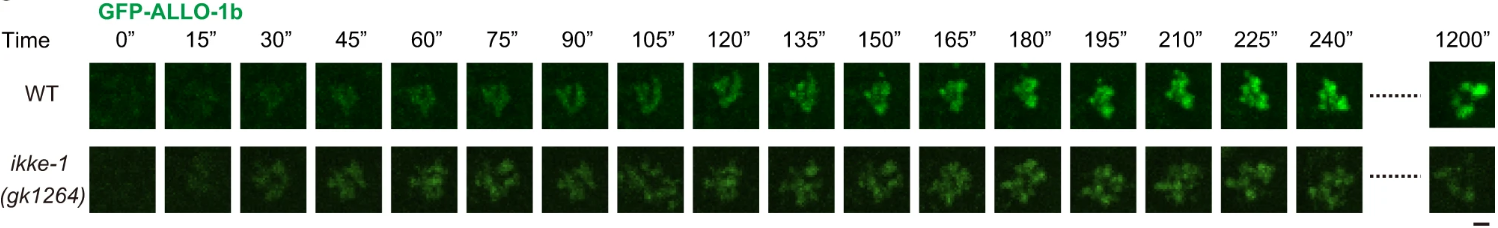
Figure 2: IKKE-1 supports the accumulation of ALLO-1b around paternal mitochondria.
The Positive Feedback Loop Driven by EPG-7
Another important element of the autophagy process uncovered by the study was a feedback mechanism involving the protein EPG-7, a homolog of the mammalian FIP200. EPG-7 plays a crucial role in maintaining ALLO-1b recruitment to paternal mitochondria, enhancing the accumulation necessary for complete autophagosome formation. The authors found that in the absence of IKKE-1, EPG-7 also fails to accumulate around paternal mitochondria. Figure 3 show the reduced intensity of EPG-7-GFP in ikke-1 mutant embryos compared to wild-type. Without sufficient EPG-7, the recruitment of other autophagic machinery is impaired, leading to incomplete autophagosome formation.
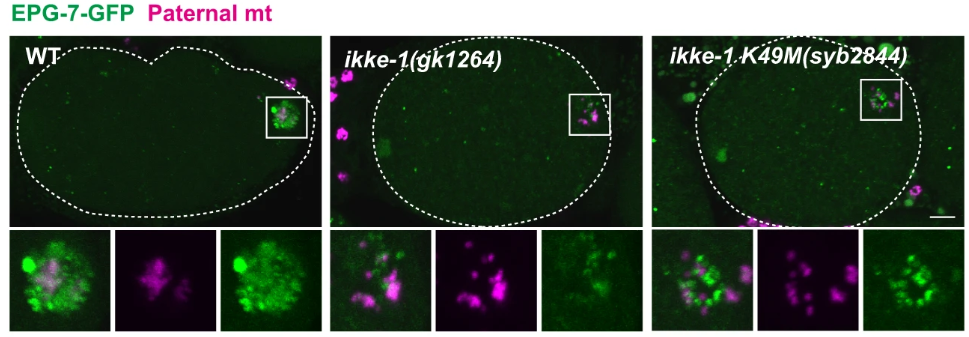
Figure 3: Reduced intensity of EPG-7-GFP in ikke-1 mutant embryos compared to wild-type.
In addition, in Figure 4, the authors demonstrate that most paternal mitochondria in ikke-1 mutants are not fully enclosed by autophagosomes, further highlighting the importance of the feedback loop in driving sustained autophagy. This feedback ensures that once ALLO-1b is recruited to mitochondria, the process continues until the organelles are fully degraded.
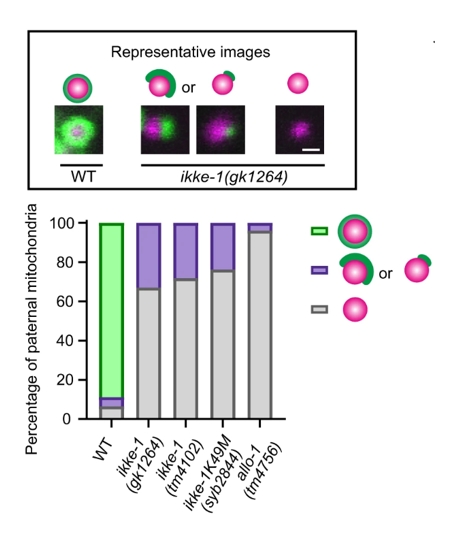
Figure 4: In ikke-1 mutants, most paternal mitochondria are not fully enclosed by autophagosomes.
IKKE-1 Phosphorylates Key Autophagic Components
Beyond just supporting ALLO-1b accumulation, IKKE-1 was found to be responsible for phosphorylating several key autophagic components. Phosphorylation is a common regulatory mechanism in cellular processes, and in this case, IKKE-1’s kinase activity was essential for modifying ALLO-1 and EPG-7 to sustain autophagosome formation. Volcano plot of tandem mass tag (TMT)-based quantitative phosphoproteo mics identifying the decrease in phosphorylation of EPG-7 S949 in ikke-1 and allo-1 mutants in Figure 5 (Red arrows indicate phosphorylated S949-containing peptides of EPG-7). It reveals that phosphorylation levels of EPG-7 were significantly reduced in ikke-1 mutants, particularly at serine residue S949. This phosphorylation appears to be a key step in stabilizing EPG-7’s interaction with ALLO-1b, as evidenced by the impaired autophagosome formation seen in these mutants.
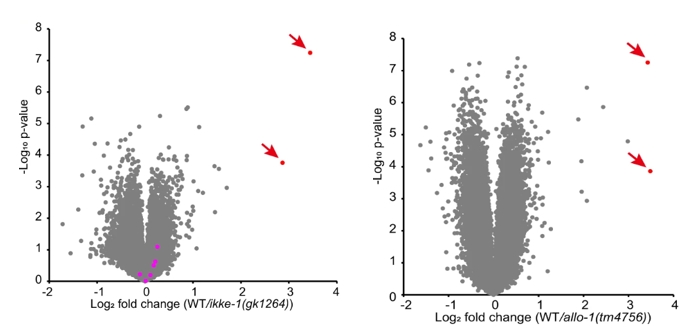
Figure 5: Volcano plot from tandem mass tag (TMT)-based quantitative phosphoproteomics identifying the decrease in phosphorylation of EPG-7 at S949 in ikke-1 and allo-1 mutants.
Conclusion:
This study provides a detailed view of how selective autophagy ensures the elimination of paternal mitochondria in Caenorhabditis elegans. By revealing the distinct roles of ALLO-1 isoforms and highlighting the essential regulatory function of IKKE-1, the researchers have uncovered a highly coordinated mechanism that maintains mitochondrial inheritance fidelity. The feedback loop(Figure 6) driven by EPG-7, coupled with IKKE-1’s phosphorylation activity, ensures that autophagosomes form efficiently around paternal mitochondria, driving their degradation.
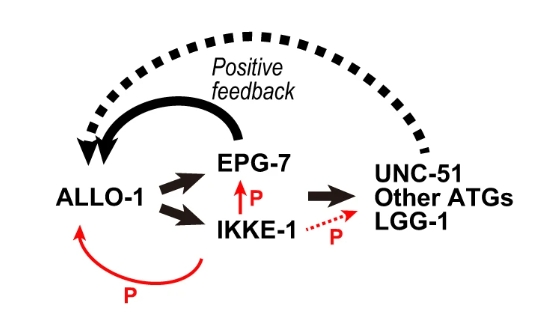
Figure 6: Model of ALLO-1 accumulation via a positive feedback loop by IKKE-1
References
Sasaki, T., Kushida, Y., Norizuki., et al. (2024). ALLO-1- and IKKE-1-dependent positive feedback mechanism promotes the initiation of paternal mitochondrial autophagy. Nature Communications, 15(1460), https://doi.org/10.1038/s41467-024-45863-2.









